Supported Applications
AlphaFold
-
Description
an implementation of the inference pipeline of AlphaFold using a completely new model that was entered in CASP14.
-
Usage
To list all executables provided by AlphaFold, run:$ biogrids-list alphafold -
Usage Notes
AlphaFold requires a set of parameters and genetic databases that must be downloaded separately. See https://github.com/deepmind/alphafold#genetic-databases for more information.
AlphaFold version 2 and version 3 have different parameters and databases and thus require different procedures.
Please see our examples:
Note that while the AlphaFold code is licensed under the open source Apache 2.0 License, the AlphaFold parameters are made available for non-commercial use only under the terms of the CC BY-NC 4.0 license. Please see the Disclaimer at https://github.com/deepmind/alphafold#license-and-disclaimer.
-
Installation
Use the following command to install this title with the CLI client:$ biogrids-cli install alphafoldAvailable operating systems: Linux 64 -
Primary Citation*
J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, and D. Hassabis. 2021. Highly accurate protein structure prediction with AlphaFold. Nature.
-
*Full citation information available through
-
-
Webinars
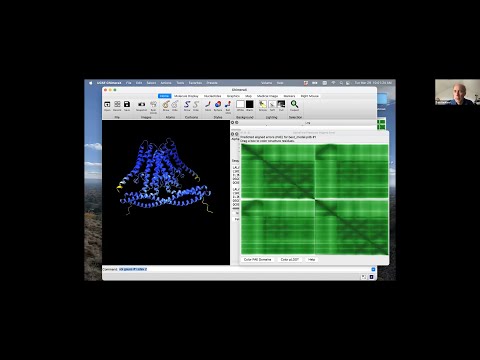
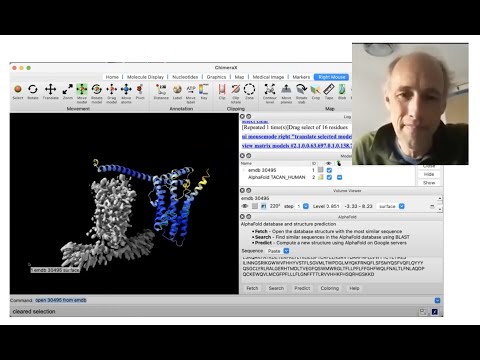
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics - Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Linked to presented materials: https://www.rbvi.ucsf.edu/chimerax/data/sbgrid-mar2022/alphafold_pae.html
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Spring Mini-series - Cryo-electron microscopy of membrane proteins: from sample to structure. See the full series lineup at https://sbgrid.org/webinars/
Organized by
Prof. Jamaine Davis, Meharry Medical College
Prof. Piotr Sliz, Harvard Medical School
Prof. Patrick Sexton, ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash University
Recorded on March 29, 2022Topic: Using AlphaFold protein structures in ChimeraX for cryoEM modeling
Presenter: Tom Goddard, UCSF Resource for Biocomputing, Visualization, and Informatics
Q&A session also includes Shaun Rawson, who presented during this session on "Effective on-the-fly and downstream processing of CryoEM data." See https://youtu.be/ra2sVFEPtN8
Learn more on the ChimeraX website:
https://www.rbvi.ucsf.edu/chimerax/data/alphafold-nov2021/af_sbgrid.html
This talk was presented as part of the SBGrid Australasian III Mini-series - CryoEM: from Sample to Structure and should appeal to both novice and expert structural biologists.
See the full mini-series lineup at https://sbgrid.org/news/sbgrid-university-otago-webinar-series.
-
Keywords
Machine Learning, Protein-Protein Interaction Prediction, Protein Structure Analysis
-
Default Versions
Linux 64: 3.0.1 (6.2 GB)
-
Other Versions
Linux 64:
2.1.2 (3.8 GB) , 2.2.0 (9.8 GB) , 2.2.2 (3.8 GB) , 2.2.3 (4.3 GB) , 2.2.4 (3.9 GB) , 2.3.0 (4.0 GB) , 2.3.1 (4.0 GB) , 2.3.2 (4.0 GB) , 2.3.2_20241024 (5.3 GB) , 3.0.0 (6.2 GB) , 3.0.0_20241202_aa724ca (6.3 GB) , 3.0.1_22b9ab8 (3.7 GB)
Developers
Tom Ward, Augustin Zidek, Saran Tunyasuvunakool, John Jumper, Demis Hassabis