A Practical Guide to NCBI BLAST

NCBI BLAST: Learn about accessing from the Entrez sequence databases; the new genome BLAST service quick finder; organism limits and other filters; formatting options and downloading options; and TreeView displays.
Presented April 29, 2016.
Get video updates, subscribe to the NCBI YouTube channel:
www.youtube.com/ncbinlm
Topic: EVcouplings: Using evolution to determine structure and function
Presenters: Kelly Brock, Ph.D., Laboratory of Debora Marks, Harvard Medical School and Thomas Hopf, Ph.D., Scientific Consultant
Host: Pete Meyer
Recorded on May 12, 2020
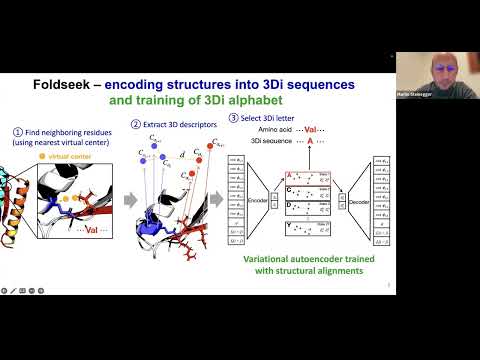
Topic: Fast and accurate protein structure search with Foldseek
Presenter: Prof. Martin Steinegger, Asst Professor of Bioinformatics, Seoul National University
Host: Pete Meyer
Recorded on Feb 14, 2023
For more information on Foldseek: https://github.com/steineggerlab/foldseek
Topic: Geneious (Custom BLAST, workflow builder, annotation tools, CRISPER tools)
Presenter: Christian Olsen, Field Application Scientist at
Biomatters, Inc
Host: Jason Key
Recorded on July 26, 2016
Topic: Geneious R7: Sequence Analysis Tool for Biologists
Presenter: Christian Olsen, Biomatters, Inc.
Host: Jason Key
Recorded on Sept 9th, 2014
Topic: Geneious R7: Molecular Cloning Tools for Biologists
Presenter: Christian Olsen, Biomatters, Inc.
Host: Jason Key
Recorded on September 30, 2014
IIHG Intro to Kallisto for RNA-Seq
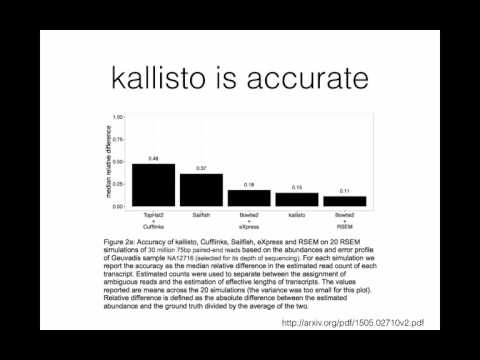
A brief introduction to the theory and method underlying kallisto for transcript quantification of RNA-Seq data.
This screencast is from our IIHG MiniCourse training series on bioinformatic methods for U Iowa researchers.
Find IIHG on the web at:
http://www.medicine.uiowa.edu/humange...
You can find me at: http://www.michaelchimenti.com; @ChimentiMichael on Twitter.
MAFFT/MAFFTash: Web services for structure-informed multiple sequence alignment
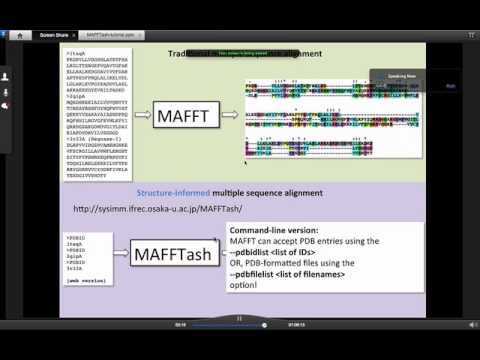
Topic: MAFFT/MAFFTash: Web services for structure-informed multiple sequence alignment
Presenters: Kazutaka Katoh and Daron Standley, Immunology Frontier Research Center, Osaka University
Host: Andrew Morin
Recorded on July 17, 2014
Molecular Biology with Geneious R7

Topic: Geneious R7: A bioinformatics platform for biologists
Presenter: Christian Olsen, Field Application Scientist, Biomatters, Inc.
SBGrid Host: Paul Sanschagrin
Recorded on Oct 3, 2013
Geneious website: http://www.geneious.com/
NCBI NOW, Lecture 7, Using BLAST for Genomic Analysis

Topic: Using BLAST for Genomic Analysis
Presenter: Tom Madden
Topic: Retraining AlphaFold2 yields new insights into its learning mechanism and capacity for generalization.
Presenter: Gustaf Ahdritz, Ph.D. Student, AlQuraishi Laboratory Columbia University.
Host: Jason Key
Recorded on April 11, 2023
For more information on OpenFold:
https://sbgrid.org/software/titles/openfold
https://github.com/aqlaboratory/openfold
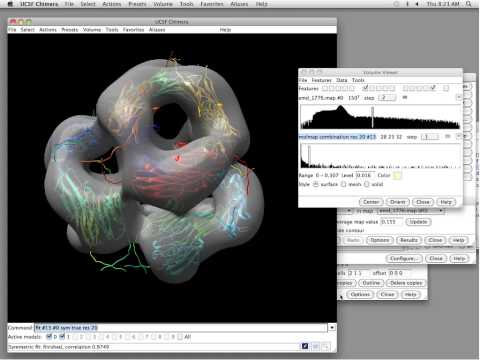
Topic: Analyzing Molecular Assemblies with UCSF Chimera"
Presenter: Tom Goddard, Programmer/Analyst, Resource for Biocomputing, Visualization, and Informatics, UCSF
Host: Jason Key
Recordeed on February 7, 2013
Using FastQC to check the quality of high throughput sequence
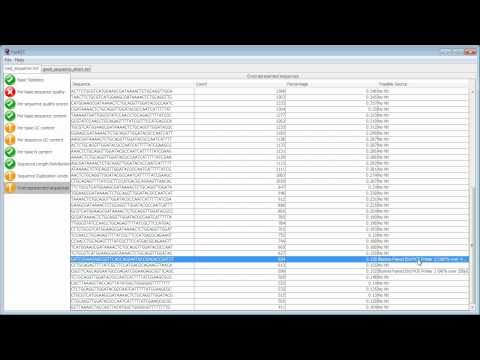
FastQC is an application which reads raw sequence data from high throughput sequencers and runs a set of quality checks to produce a report which allows you to quickly assess the overall quality of your run, and to spot any potential problems or biases.
This video demonstrates the use of FastQC to analyse some sequence data and goes through the results to explain what a good dataset looks like, and what sort of problems you might encounter.
Using IGV Browser for Variants and Next Generation Sequencing (Part 1)
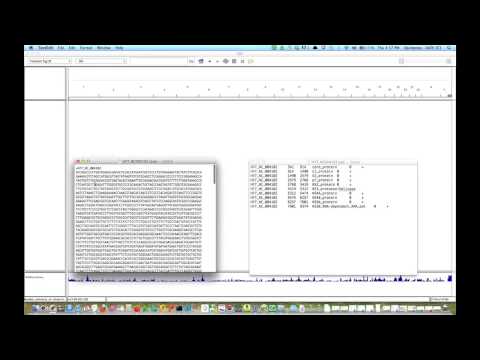
This video demonstrates how to use IGV browser for a custom genome and dataset, how to use the launch tool, how to import a genome and annotations, and how to import a dataset. Also shown: how to navigate, search, and annotate regions of interest.














