Webinars
- Page 1 of 1

NCBI BLAST: Learn about accessing from the Entrez sequence databases; the new genome BLAST service quick finder; organism limits and other filters; formatting options and downloading options; and TreeView displays.
Presented April 29, 2016.
Get video updates, subscribe to the NCBI YouTube channel:
www.youtube.com/ncbinlm
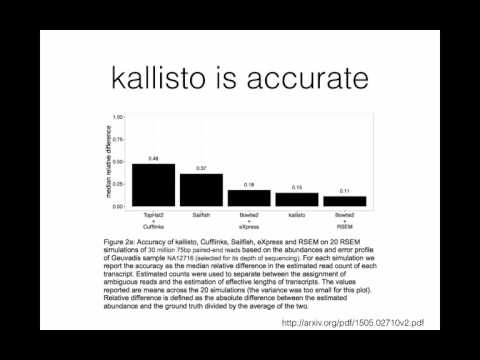
A brief introduction to the theory and method underlying kallisto for transcript quantification of RNA-Seq data.
This screencast is from our IIHG MiniCourse training series on bioinformatic methods for U Iowa researchers.
Find IIHG on the web at:
http://www.medicine.uiowa.edu/humange...
You can find me at: http://www.michaelchimenti.com; @ChimentiMichael on Twitter.
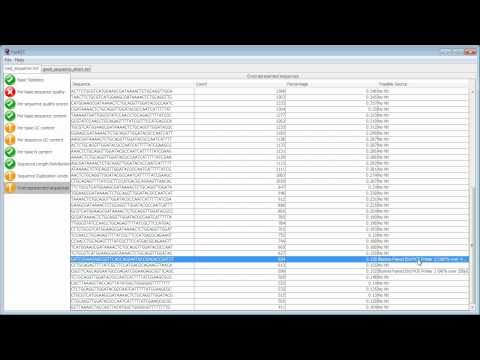
FastQC is an application which reads raw sequence data from high throughput sequencers and runs a set of quality checks to produce a report which allows you to quickly assess the overall quality of your run, and to spot any potential problems or biases.
This video demonstrates the use of FastQC to analyse some sequence data and goes through the results to explain what a good dataset looks like, and what sort of problems you might encounter.

