Pipeline Dreams
Bridget Carragher and Clint Potter
Simons Electron Microscopy Center at the New York Structural Biology Center
Published April 26, 2016
A year ago, Bridget Carragher and Clint Potter’s group broke the so-called three-angstrom barrier for electron microscopy (EM). Prior to their work, so many structures had been solved using EM at 3.4 or 3.5-angstrom resolution that people had started to believe higher resolutions were out of reach with the technology.
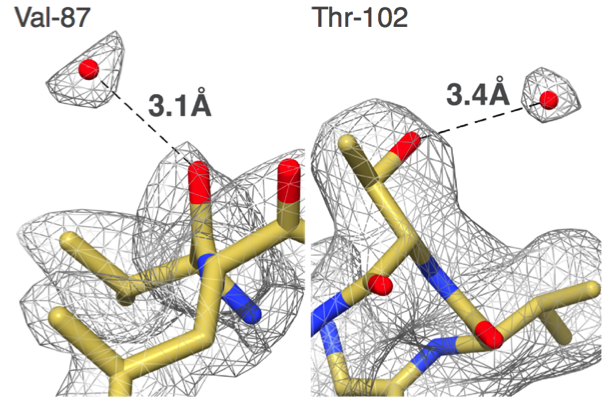
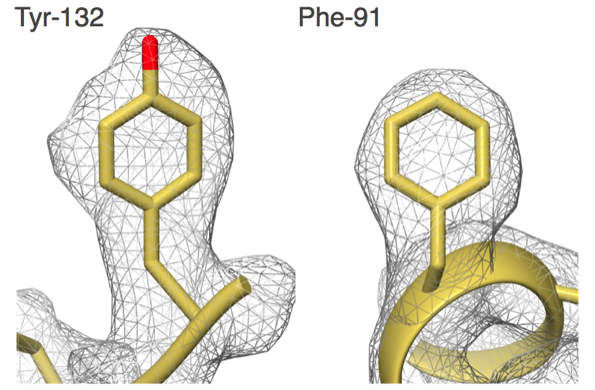
"Our group set out to show that EM could do better,” says Carragher, co-Director of the Simons Electron Microscopy Center (SEMC) at the New York Structural Biology Center (NYSBC). “We did things carefully and in a fully automated fashion and got to 2.8 angstroms. That was the first time anyone was able to see water molecules in an EM structure."
What was a big deal then has become commonplace now, but only because the EM field is changing so rapidly. Carragher and Clint Potter, the other SEMC co-Director, are among the drivers in the field with their work on EM automation tools such as Leginon and Appion.
Their journey in this field began about twenty years ago when they met at the University of Illinois at the Beckman Institute. Carragher was working on EM, and Potter on magnetic resonance imaging. But they were both interested in the automated control of instruments. In the late 1990s, they wrote their first software tools for controlling electron microscopes, among which was an early incarnation of Leginon.
The need for automated tools such as Leginon developed when EM switched from film to digital images. The field of view of digital images is smaller than film, so more need to be taken to get the same amount of information. Instead of hundreds of images, a researcher might need to collect thousands. Graduate students have been known to spend three or four days collecting images even with automation tools at their disposal. “Once you go digital, you have to automate,” says Carragher. “To me there’s no real choice because it’s so tedious.”
In 2001, the Scripps Research Institute recruited Carragher and Potter. Soon after, they received a grant that allowed them to work full-time on automation tools. Their first task was to rewrite Leginon to turn it into professional software that could be maintained and expanded. Before the rewrite, Leginon, says Carragher, “had been hacked together with a bunch of scripts written in Tkl/Tk and some other some bizarre packages.”
They spent a year on the overhaul, this time using Python. During that year, they had nothing. At times, it felt like progress had come to a halt. And in the end, they had the same features that they started with. “Scary times,” says Potter.
Carragher and Potter’s group also developed an automated tool called Appion to process the data produced by Leginon, creating an automation pipeline for single particle EM. Another reason so many images must be taken to get a clear structure with high resolution is that each individual image is grainy since only a few electrons can be bounced off of a sample before destroying it. Those images flow into Appion, which lines them up and processes them.
About ten years ago, Appion was processing data from about 250,000 particles. Today, researchers are using a million particles to get clearer images and a better sense of heterogeneity. “If all the particles are the same, your job is simple,” says Carragher. “But molecular machines found in nature have moving parts. You need to sort that out to tell, for example, if the machine is in a closed or open state.”

In 2015, Carragher and Potter relocated from Scripps to the New York Structural Biology Center to set up the Simons Electron Microscopy Center using funding from the Simons Foundation. The National Resource for Automated Molecular Microscopy moved to SEMC with them. SEMC is still bringing in new equipment—a new Titan Krios has just been accepted and a second one will be delivered in August 2016—but the Center is up and running and already supporting about a hundred different projects. “We’re serving this enormous group of people and are upgrading this facility to soon — I hope — be one of the best in the world,” says Carragher.
While Leginon and Appion are mature, there is a constant need to support new technology, such as Phase Plates for Cryo-transmission electron microscopy (TEM) and other new devices. Further, the team is working on advancing specimen preparation hardware.
A new automation project is also underway to build an automated pipeline similar to Leginon and Appion, but that spans scales and technologies and can provide a means to look at very large structures, such as a thin slice of a cell. The team envisions integration across scales by integrating images from different sources, such as light microscopes, focused ion beam scanning electron microscopes (FIBSEM), and TEM.
The SEMC team Carragher and Potter have formed includes a group of professional software engineers and staff scientists. This group is critical for achieving continuity. “Nobody wants to have to continuously take over codes from a postdoc,” says Potter. “We’ve been successful with a core group that keeps the software going.”
The SEMC team also includes graduate students and postdocs, who drive the technology by doing challenging biology and also often make major contributions to the technology and software. These young scientists cross-embed themselves at both SEMC and in biology labs to form productive and inventive collaborations. “The biology is driven by a proper biology lab, but the technology is driven here,” says Carragher.
“There’s synergy between the labs,” adds Potter.
“And,” Carragher says, “It lets us tackle some beautifully interesting projects.”
--Elizabeth Dougherty