Computing Cellular Clockworks
Klaus Schulten
University of Illinois, Urbana-Champaign
Published October 23, 2015
Physicist Klaus Schulten once imagined becoming a dancer, relying on nothing but his own mind and body to perform. “But I was not a good dancer,” he says. “So my next thing was theoretical physicist. Just myself, pencil and paper — and in my case, also an eraser.”
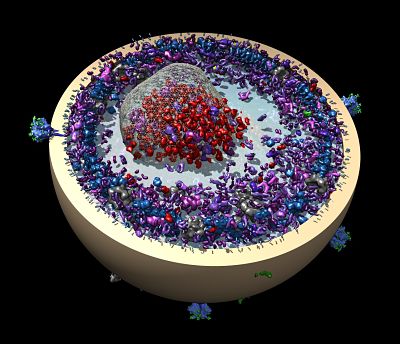
That dream was also thwarted. Today, Schulten relies on some of the most powerful and expensive computing equipment on earth to carry out his work, which applies theoretical physics to the understanding of biological systems. His most recent work involved the molecular simulation of an organelle that converts light energy to chemical energy inside a photosynthetic bacterium using the molecular dynamics tools NAMD and VMD, which his lab develops. Looking forward, he wants to simulate an entire living cell, right down to the last atom. “If we want to understand health and disease, we need to understand life at the molecular level and to know how all the molecular components work together like clockwork,” says Schulten, the Swanlund Professor of Physics at the University of Illinois at Urbana-Champaign.
As an undergraduate in Germany in the 1960s, Schulten chose to study physics because he was good at mathematics, but interested in biology. “I thought physics was in the middle,” he says.
When choosing a graduate school, he sought out Harvard University’s Martin Karplus because he was one of the few people blending theoretical physics and chemistry with biology. With Karplus, Schulten studied vision, working to understand how biomolecules absorb light and change as a result. Later, in 2013, Karplus won a Nobel Prize for molecular dynamics methods in chemistry.
Afterwards, in 1976, Schulten returned to Germany to work in the vicinity of another Nobel Laureate, Manfred Eigen, at the Max Planck Institute for Biophysical Chemistry. “By the name, it was clearly an interdisciplinary institute,” says Schulten.
There, Schulten began his work on electron transfer reactions, which are critical for the conversion of sunlight to the chemical energy needed for living cells. This work marks the beginnings of what became a forty-year effort to understand photosynthesis at the atomic and even electronic level. Schulten made interesting and surprising discoveries, such as the fact that electron transfer reactions are affected by magnetic fields.
He also established his modus operandi. As one of a handful of theoreticians among hundreds of experimentalists at Max Planck, Schulten was one of the go-to guys for experimentalists with puzzling results. “I worked on interesting questions that begged for an answer, and I answered by bringing together biology, physics and chemistry,” he says.
Soon, however, he recognized the limits of pencil and paper. “If I wanted to do real biology, I needed to do computing,” he says.Schulten considered purchasing computing equipment, but what he could afford would not provide the computing power he needed. So he and his students built their own parallel computer. For about $30,000, their machine had the power of a Cray-2. Instead of being awed by this feat, his colleagues in physics looked down their noses at computational work, as if it was a sign of intellectual weakness. “They all wanted to be genius Einsteins,” he says. “I myself wanted to study living systems from the perspective of physics.”
So in the 1980s, Schulten moved, first to the Technical University of Munich, and then to the newly formed Beckman Institute at the University of Illinois. Schulten was the first to move in. There he formed, with support of the National Institutes of Health and National Science Foundation, the Theoretical and Computational Biophysics Group, which today has approximately 35 members, including physicists, chemists, computer scientists, biophysicists, computer programmers, and many graduate students.
At Beckman, Schulten enhanced his molecular dynamics simulator. The original code had been based on an obscure language, OCCAM, attached to the Transputer chipset he’d used in his lab-made parallel computer. His lab members rewrote the program in C, a bold step at the time, since most scientists were using Fortran.
In the early 1990s, Schulten’s students revolted, demanding yet another fresh start. “I said do it,” he says. “I really like initiative.”
The students wrote a completely new program, this time in C++, called NAMD. The language was very new at the time, and again, Schulten faced ridicule from colleagues. But he forged ahead, not only using the new language, but also taking advantage of its structured approach to build parallel processing into the software. His lab also decided to make the software free and open.
The earliest NAMD simulations involved about thirty thousand atoms, but the numbers grew to a hundred thousand, then a million. Today, the largest simulations include a hundred million atoms. This growth in scale inspired the development of VMD, a sister program for NAMD for visualizing the simulations. Today, NAMD has about ten thousand users, VMD, several hundred thousand.
In 2015, Schulten used NAMD and VMD to simulate and visualize the structure and function of a bacteria’s photosynthetic engine, called the chromatophore. The chromatophore is a sphere-shaped organelle, approximately one one-hundredth of the cell’s total volume, consisting of 100 million atoms, 16,000 lipids, and 101 proteins. The team performed the simulation on the Titan supercomputer at the US Department of Energy’s Oak Ridge National Laboratory in Tennessee. “It’s the biggest simulation we’ve done,” says Schulten.
For Schulten, this project is just another step in what has been a decades-long journey for his lab and also for structural biology. Much work has gone into simulating single proteins and complexes, but that’s just the beginning. “The body is an aggregate, a very hierarchical aggregate of biomolecules,” he says. “What people want to see now is how these molecules aggregate and form teams and factories working together.”
The next logical step after simulating an organelle is to simulate an even bigger aggregate: the cell. “The cell is very stable,” he says. “It acts as quite a precise clockwork despite all the thermal noise characteristic for molecules acting at physiologic temperature. We want to understand the clockwork.”
The idea of simulating an entire living cell may seem impossible. But Schulten is used to skeptics. He is also used to doing things no one has ever done before. In fact, he has the steps ahead already mapped out.
Current computing power allows for the simulation of approximately one one-hundredth of a small cell, as demonstrated by the photosynthetic chromatophor project. Schulten reasons that simulating an entire small bacterial cell will require 100-times the current computing power. That power — provided by an exoscale computer — may be within reach. By 2018, Oak Ridge plans to complete the Summit computer, which will be 10-times more powerful than the 10-petaflop Titan. Projections put a 1000-petaflop exoscale computer’s availability around 2022. Schulten’s lab is already working with Oak Ridge and IBM to build software that makes efficient use of the Summit computer. “We are doing everything we can to work towards the simulation of a whole living cell,” he says. “Getting there will offer many exciting milestones in computational cell biology. The path is often as exciting as the final goal.”
-- Elizabeth Dougherty